Molecular genetic analyses of 300-year old skeletons from Auersperg tomb
Abstract
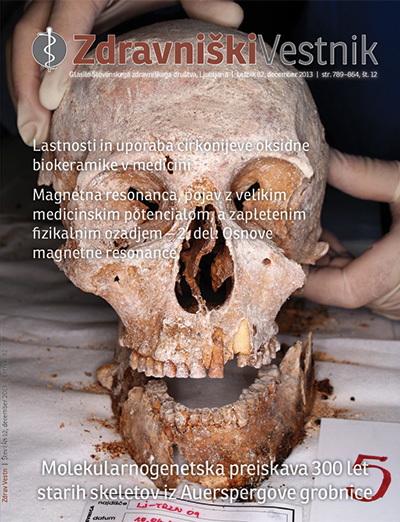
Background: In 2009 the archaeologists excavated five skeletons from a 17th-century archaeological site in Ljubljana. They were found in the side chapel of the church in the Franciscans monastery, which was the Auerspergs’ tomb. Beside the skeletons, the finds revealed a bronze bowl with the heart , and the name of Ferdinand II and the years of birth and death (1655–1706) engraved. In 2011, we were asked to identify those five skeletons. The skeletons were poorly preserved and bones degraded to small pieces. Fragments of femurs and teeth were preserved only in two skeletons, therefore for the remaining three the fragments of cranium were used for molecular genetic analyses.
Methods: We cleaned the bones and teeth, removed surface contamination, and ground them into powder. Prior to DNA isolation, bone or tooth powder was decalcified. DNA was purified in the Biorobot EZ1 device (Qiagen). Nuclear DNA of the samples was quantified using real-time polymerase chain reaction (PCR). Short tandem repeat (STR) typing of autosomal DNA was performed using Investigator ESSplex Kit (Qiagen), the NGM Kit (Applied Biosystems) and the MiniFiler Kit (Applied Biosystems). Typing of the Y-STRs was performed using the YFiler Kit (Applied Biosystems). The two hypervariable regions HVI and HVII of the mtDNA were sequenced.
Results: We were able to extract up to 10.7 ng DNA/g of tooth powder from Auersperg chapel archaeological site skeletal remains. We managed to obtain nuclear DNA for successful STR typing from skeletal remains that were over 300 years old. From one skeleton we obtained a complete male genetic profile of autosomal DNA, almost complete Y-STR haplotype, which enabled us to track the paternal line and mtDNA haplotype for HVI and HVII regions, which enabled us to track the maternal line. After comparing the profiles with elimination database, no match was found, and thus the authenticity of genetic profiles was confirmed.
Conclusions: Now we are waiting for the family reference samples for comparison with genetic profiles obtained in order to identify the excavated skeletons. This is the first archaeogenetic research performed in Slovenia.
Downloads
References
Kemp BM, Smith DG. Use of bleach to eliminate contaminating DNA from the surface of bones and teeth. Forensic Sci Int 2005; 154: 53–61.
Hofreiter M, Serre D, Poinar HN, Kuch M, Pääbo S. Ancient DNA. Nat Rev Genet 2001; 2: 353–59.
Alaeddini R, Walsh SJ, Abbas A. Forensic implications of genetic analysis from degraded DNA–A review. Forensic Sci Int Genet 2010; 4: 149–157.
Alaeddini R. Forensic implication of PCR inhibition–A review. Forensic Sci Int Genet 2012; 6: 297–305.
Anderung C, Persson P, Bouwman A, Elburg R, Gotherstrom A. Fishing for ancient DNA. Forensic Sci Int Genet 2008; 2: 104–107.
Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res 1996; 24: 1304–1307.
Poinar HN, Hoss M, Bada JL, Pääbo S. Amino acid racemisation and the preservation of ancient DNA. Science 1996; 272: 864–866.
Lindahl T. Instability and decay of the primary structure of DNA. Nature 1993; 362: 709–15.
Smith CI, Chamberlain AT, Riley MS, Cooper A, Stringer CB, Collins MJ. Neanderthal DNA. Not just old but old and cold? Nature 2001; 410: 771–772.
Smith CI, Chamberlain AT, Riley MS, Stringer C, Collins MJ. The thermal history of human fossils and the likelihood of successful DNA amplification. J Hum Evol 2003; 45: 203–217.
Burger J, Hummel S, Hermann B, Henke W. DNA preservation: a microsatellite-DNA study on ancient skeletal remains. Electrophoresis 1999; 20: 1722–28.
Pruvost M, Schwarz R, Correia VB. Freshly excavated fossil bones are best for amplification of ancient DNA. Proc Natl Acad Sci USA 2007; 104: 739–744.
Putkonen MT, Palo JU, Cano JM, Hedman M, Sajantila A. Factors affecting the STR amplification sucess in poorly preserved bone samples. Investigative genetics 2010; 1: 9.
Irwin JA, Just RS, Loreille OM, Parsons TJ. Characterization of a modified amplification approach for improved STR recovery from severely degraded skeletal elements. Forensic Sci Int Genet 2012; 6: 578–587.
Jakubowska J, Maciejewska A, Pawlowski R. Comparison of three methods of DNA extraction from human bones with different degrees of degradation. Int J Legal Med 2012; 126: 173–178.
Loreille OM, Diegoli TM, Irwin JA, Coble MD, Parsons TJ. High efficiency DNA extraction from bone by total demineralization. Forensic Sci Int Genet 2007; 1: 191–5.
Amory S, Huel R, Bilić A, Loreille O, Parsons TJ. Automatable full demineralization DNA extraction procedure from degraded skeletal remains. Forensic Sci Int Genet 2012; 6: 398–406.
Zupanič Pajnič I. Visoko učinkovita metoda ekstrakcije DNA iz skeletnih ostankov (Highly efficient DNA extraction method from skeletal remains). Zdrav Vestn 2011; 80: 171–181.
Zupanič Pajnič I, Balažic J, Komel R. Sequence polymorphism of the mitochondrial DNA control region in the Slovenian population. Int J Legal Med 2004; 118: 1–4.
Šterlinko H, Zupanič Pajnič I, Balažic J, Komel R. Human Y-specific STR haplotypes in a Slovenian population sample. Forensic Sci Int 2001; 120: 226–228.
Gusmao L, Butler JM, Carracedo A, Gill P, Kayser M, Mayr WR, Morling N, Prinz M, Roewer L, Tyler-Smith C, Schneider PM. DNA commission of the international society of forensic genetics (ISFG): an update of the recommendations on the use of Y STRs in the forensic analysis. Int J Legal Med 2006; 120: 191–200.
Palo JU, Hedman M, Soderholm N, Sajantila A. Repatriation and identification of Finnish World War II soldiers. Croat Med J 2007; 48: 528–535.
Jehaes E, Decorte R, Peneau A, Petrie JH, Boiry PA, Gilissen A, et al. Mitochondrial DNA analysis on remains of a putative son of Louis XVI king of France and Marie-Antoinette. Eur J Hum Genet 1998; 6: 383–95.
Jehaes E, Toprak K, Vanderheyden N, Pfeiffer H, Cassiman JJ, Brinkmann B, et al. Pitfalls in the analysis of mitochondrial DNA from ancient specimens and the consequences for forensic DNA analysis: the historical case of the putative heart of Louis XVII. Int J Legal Med 2001; 115: 135–41.
Anslinger K, Weichhold G, Keil W, Bayer B, Eisenmenger W. Identification of the skeletal remains of Martin Bormann by mtDNA analysis. Int J Legal Med 2001; 114: 194–196.
Stone AC, Starrs JE, Stoneking M. Mitochondrial DNA analysis of the presumptive remains of Jesse James. J Forensic Sci 2001; 46: 173–176.
Irwin JA, Leney MD, Loreille O, Barritt SM, Christensen AF, Holland TD, Smith BC, Parsons TJ. Application of low copy number STR typing to the identification of aged, degraded skeletal remains. J Forensic Sci 2007; 52: 1322–1327.
Zupanič Pajnič I. Molekularnogenetska identifikacija domobranskih žrtev (Molecular genetic identification of Slovenian home guard victims). Zdrav Vestn 2008; 77: 745–750.
Zupanič Pajnič I, Gornjak Pogorelc B, Balažic J. Molecular genetic identification of skeletal remains from the Second World War Konfin I mass grave in Slovenia. Int J Legal Med 2010; 124: 307–317.
Bogdanowicz W, Allen M, Branicki W, Lembring M, Gajewska M, Kupiec T. Genetic identification of putative remains of the famous astronomer Nicolaus Copernicus. Proc Natl Acad Sci USA 2009; 106: 12279–82.
Kupiec T, Branicki W. Genetic examination of the putative skull of Jan Kochanowski reveals its female sex. Croat Med J 2011; 52: 403–409.
Graham EAM. DNA reviews: Ancient DNA. Forensic Sci Med Pathol 2007; 3: 221–225.
Zupanič Pajnič I. Priporočila za molekularno genetsko identifikacijo žrtev povojnih pobojev v Sloveniji. V: Dežman J, ur. Poročilo Komisije vlade Republike Slovenije za reševanje vprašanj prikritih grobišč 2005–2008. Ljubljana: Družina; 2008. p. 133–146.
Pääbo S, Poinar H, Serre D. Genetic analyses from ancient DNA. Annu Rev Genet 2004; 38: 645–79.
Edson SM, Ross JP, Coble MD, Parsons TJ, Barritt SM. Naming the dead–confronting the realities of rapid identification of degraded skeletal remains. Forensic Sci Reviews 2004; 16: 64–89.
Zupanič Pajnič I, Gornjak Pogorelc B, Balažic J, Zupanc T, Štefanič B. Highly efficient nuclear DNA typing of the World War II skeletal remains using three new autosomal short tandem repeat amplification kits with the extended European Standard Set of loci. Croat Med J 2012; 53: 17–23.
Miloš A, Selmanović A, Smajlović L, Huel RLM, Katzmarzyk C, Rizvić A, Parsons JP. Success rates of nuclear short tandem repeat typing from different skeletal elements. Croat Med J 2007; 48: 486–493.
Misner LM, Halvorson AC, Dreier JL, Ubelaker DH, Foran DR. The correlation between skeletal weathering and DNA quality and quantity. J Forensic Sci 2009; 54: 822–828.
Davoren J, Vanek D, Konjhodzić R, Crews J, Huffine E, Parsons TJ. Highly effective DNA extraction method for nuclear short tandem repeat testing of skeletal remains from mass graves. Croat Med J 2007; 48: 478–85.
Alonso A, Andelinović Š, Martin P. DNA typing from skeletal remains: evaluation of multiplex and megaplex STR systems on DNA isolated from bone and teeth samples. Croat Med J 2001; 42: 260–6.
Tully G, Bär W, Brinkmann B, Carracedo A, Gill P, Morling N. Considerations by the European DNA profiling (EDNAP) group on the working practices nomenclature and interpretation of mitochondrial DNA profiles. Forensic Sci Int 2001; 124: 83–91.
Bär W, Brinkmann B, Budowle B, Carracedo A, Gill P, Holland M. DNA commission of the International society for forensic genetics: guidelines for mitochondrial DNA typing. Int J Legal Med 2000; 113: 193–196.
Carracedo A, Bär W, Lincoln P, Mary W. DNA commission of the International society for forensic genetics: guidelines for mitochondrial DNA typing. Forensic Sci Int 2000; 110: 79–85.
Wilson MR, DiZinno JA, Polanskey D, Replogle J, Budowle B. Validation of mitochondrial DNA sequencing for forensic casework analysis. Int J Legal Med 1995; 108: 68–74.
Kalmar T, Bachrati CZ, Marcsik A, Rasko I. A simple and efficient method for PCR amplifiable DNA extraction from ancient bones. Nucleic Acids Res 2000; 28: 67.
Tamariz J, Voynarovska K, Prinz M, Caragine T. The application of ultraviolet irradiation to exogenous sources of DNA in plasticware and water for the amplification of low copy number DNA. J Forensic Sci 2006; 51: 790–794.
Shaw K, Sesardić I, Bristol N, Ames C, Dagnall K, Ellis C, et al. Comparison of the effects of sterilisation techniques on subsequent DNA profiling. Int J Legal Med 2008; 122: 29–33.
Vanek D, Saskova L, Koch H. Kinship and Y-chromosome analysis of 7th century human remains: Novel DNA extraction and typing procedure for ancient material. Croat Med J 2009; 50: 286–295.
Rohland N, Hofreiter M. Ancient DNA extraction from bones and teeth. Nature Protocols 2007; 2: 1756–1762.
Qiagen Companies, EZ1 DNA Handbook, Vienna, 2004.
Qiagen Companies, TissueLyser Handbook, Vienna, 2004.
Applied Biosystems, Quantifiler™ human DNA quantification kit user guide, Foster City, 2003.
Gross AM, Liberty AA, Ulland MM, Kuriger JK. Internal validation of the AmpFlSTR Yfiler amplification kit for use in forensic casework. J Forensic Sci 2008; 53: 1–10.
Applied Biosystems, AmpFlSTR® NGM™ PCR amplification kit user guide, Foster City, 2009.
Qiagen Companies, Investigator ESSplex handbook, Vienna, 2010.
Applied Biosystems, AmpFlSTR MiniFiler™ PCR Amplification kit user guide, Foster City, 2007.
Applied Biosystems, AmpFlSTR YFiler™ PCR Amplification kit user guide, Foster City, 2005.
Parson W, Parsons TJ, Scheithauer R, Holland MM. Population data for 101 Austrian Caucasian mitochondrial DNA d-loop sequences: Application of mtDNA sequence analysis to a forensic case. Int J Legal Med 1998; 111: 124–132.
http://www.mbio.ncsu.edu/BioEdit/bioedit.htm
Zupanc T, Štefanič B, Balažic J and Zupanič Pajnič I. Performance of the Human Quantifiler, the Investigator Quantiplex, and the Investigator ESSplex Plus kit for quantification and nuclear DNA typing of old skeletal remains. Rom J Legal Med 2013; in press.
Montpetit SA, Fitch IT, O`Donnell PT. A simple automated instrument for DNA extraction in forensic casework. J Forensic Sci 2005; 50: 1–9.
Valgren C, Wester S, Hansson O. A comparison of three automated DNA purification methods in forensic casework. Forensic Sci Int: Genetics Supplement Series 2008; 1: 76–77.
Kishore R, Hardy WR, Anderson VJ, Sanchez NA, Buoncristiani MR. Optimization of DNA extraction from low-yield and degraded samples using the biorobot EZ1 and biorobot M48. J Forensic Sci 2006; 51: 1055–61.
Zupanič Pajnič I. A comparative analysis of the AmpFlSTR Identifiler and PowerPlex 16 autosomal Short Tandem Repeat (STR) amplification kits on the skeletal remains excavated from Second World War mass graves in Slovenia. Rom J Legal Med 2013; 21(1): 73–78.

The Author transfers to the Publisher (Zdravniški vestnik/Slovenian Medical Journal) all economic copyrights following form Article 22 of the Slovene Copyright and Related Rights Act (ZASP), including the right of reproduction, the right of distribution, the rental right, the right of public performance, the right of public transmission, the right of public communication by means of phonograms and videograms, the right of public presentation, the right of broadcasting, the right of rebroadcasting, the right of secondary broadcasting, the right of communication to the public, the right of transformation, the right of audiovisual adaptation and all other rights of the author according to ZASP.
The aforementioned rights are transferred non-exclusively, for an unlimited number of editions, for the term of the statutory
The Author can make use of his work himself or transfer subjective rights to others only after 3 months from date of first publishing in the journal Zdravniški vestnik/Slovenian Medical Journal.
The Publisher (Zdravniški vestnik/Slovenian Medical Journal) has the right to transfer the rights, acquired parties without explicit consent of the Author.
The Author consents that the Article be published under the Creative Commons BY-NC 4.0 (attribution-non-commercial) or comparable licence.